Spatial Transcriptome Services
Spatial Transcriptome Gene Expression;
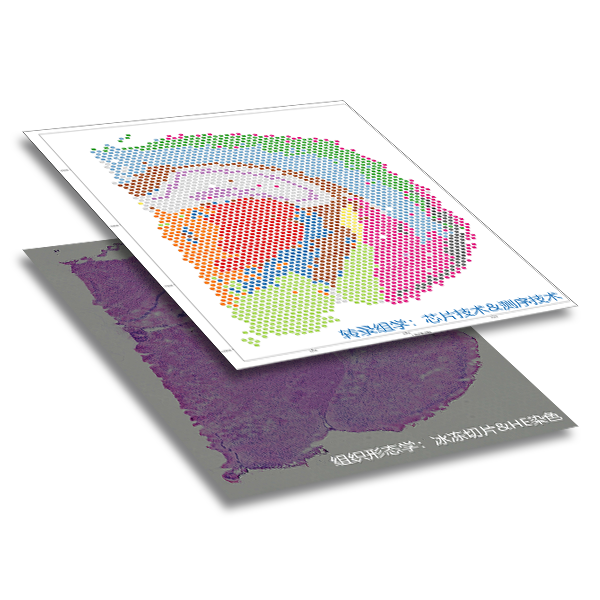
Combining histology (frozen section &HE (hematoxylin eosin staining) with transcriptomics (chip technology & sequencing technology) : deciphering spatial localization of gene expression.
Introduction
Spatial transcriptome:
A technique for detecting transcriptomic gene expression in situ in tissues, integrating the gene expression with the original tissue slice staining image. The spatial location information of gene expression in tissues can be obtained while detecting gene expression level.
Why spatial transcriptome?
Gene expression is spatiotemporal, and time-specific can be analyzed by using sequencing technology to analyze cell types and gene expression patterns in time dimensions through sampling samples at different time points. Spatially specific information is relatively difficult to obtain.
As the basic unit of organisms, cells do not exist independently, and their spatial location is the key factor to determine their properties, such as tumor microenvironment and the development of three germ layers of gastrula.
Both bulk RNA-seq and single-cell RNA-seq sequencing are difficult to restore the original location information of cells.
Sm-FISH is difficult to achieve high throughput detection.
Features
1. Obtain high-throughput gene expression analysis in multiple sample slices;
2. Reveal the biological structure of normal and pathological tissue samples;
3. There is no bias introduced by amplification and the mRNA information is captured in situ;
4. Trace the transcriptome sequencing data back to the original tissue morphology;
5. Close to single-cell resolution;
6. No need to dissociate the sample;
7. Discover new organizational markers.
Parameters
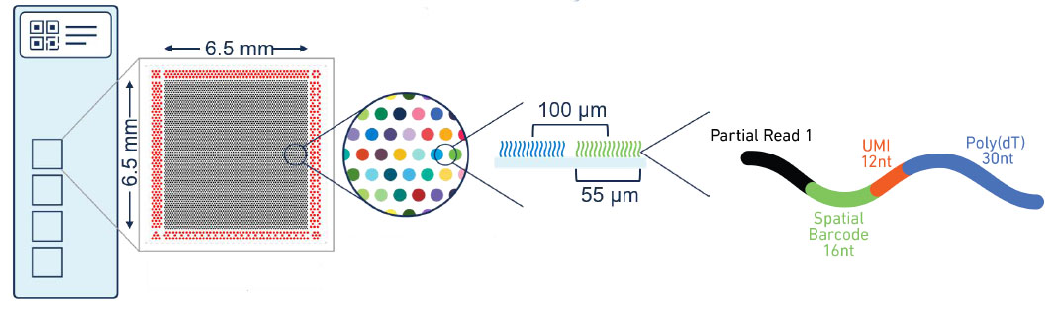
Visium spatial transcriptome gene expression chip
The chip consists of four capture regions, each of which contains about 5,000 spatial position points labeled with physical codes, and each point contains millions of capture oligonucleotide probes with spatial barcodes. After release, tissue mRNA combines with oligonucleotides with barcodes to capture gene expression information.
Capture area specification:6.5*6.5mm;
Spot diameter:55μm;
Center distance between spots:100μm;




